Abschlussarbeiten
Themen für Abschlussarbeiten ergeben sich auch aus den laufenden Forschungsprojekten am Fachgebiet. Bei Interesse empfehlen wir Ihnen, sich direkt in der Sprechstunde über derzeit verfügbare Themen zu informieren. Nach Absprache mit der/dem Betreuer*in können die Abschlussarbeiten in Deutsch oder Englisch erstellt werden.
Angebotene Abschlussarbeiten
Aktuelle Themen für Bachelor-, Master-, Studien- und Diplomarbeiten finden sich hier. Eigene Themenvorschläge sind ebenso willkommen.
- Vergleichende Genomanalyse von Eisen- und Mangan präzipitierenden Bakterien (MSc)
Untersuchungen zur Biofilmdetektion und deren Vermeidung im Waschgerät (MSc)
Abgeschlossene Arbeiten
2022
- Characterization of a coliphage isolated from the Erpe River and its potential fate in the riverine hyporheic zone.
2021
- Strategien zur Kultivierung und Untersuchung der Biofilme von Aquabacterium citratiphilum auf Mikroplastikpartikeln
- Analyse von Mangan-oxidierenden Bakteriengemeinschaften aus Manganfestbett-Bioreaktoren und Untersuchungen zum Abbau von Schadstoffen durch Mangan-oxidierende Bakterien
- Biotransformation von Benzotriazol und Metoprolol durch Mangan-oxidierende Bakterien
- Investigating the role of biological manganese oxidation in the removal of microcystin
- Isolation and Characterization of Bacteriophages Infecting Bacillus spizizenii DSM 347, Clostridium bornimense sp. nov. M3/6T, and Proteiniphilum saccharofermentans sp.nov. M2/40T from Biogas Reactors.
- Phage therapy of pulmonary Pseudomonas aeruginosa infections in cystic fibrosis patients: success of phage therapy in dependency of metabolic state of the host bacteria.
- Isolation and characterization of phages infecting the Comamonadaceae bacterium Ideonella sp. A288.
2020
- Development of a Rapid Test for Biofilm Quantification in Household Appliances
- Untersuchung zur Rolle mikrobieller Prozesse bei der Entfernung von Spurenstoffen und ihren oxidierten Transformationsprodukten in Anthrazit- und Aktivkohle-Festbettreaktoren
2019
- Analyse der Biofilmbildung in handelsüblichen Waschmaschinen
- Einfluss von Manganoxiden und Biochips auf das Wachstum von Biofilmen und den Abbau nvon Pharmaka in Bioreaktoren
- Optimisation of the Real-Time Polymerase Chain Reaction (qPCR) Method for the Quantification of Bacteriophages in Environmental Samples
- Abbau von Pharmaka durch Biomasse aus Rückspülschlamm von Enteisenungs- und Entmanganungsfiltern eines Wasserwerks
- Optimisation of the Real-Time Polymerase Chain Reaction (qPCR) Method for the Quantification of Bacteriophages in Environmental Samples.
2018
- Analyse der Biofilmbildung in handelsüblichen Waschmaschinen
- Einfluss von Manganoxiden und Biochips auf das Wachstum von Biofilmen und den Abbau nvon Pharmaka in Bioreaktoren
- Optimisation of the Real-Time Polymerase Chain Reaction (qPCR) Method for the Quantification of Bacteriophages in Environmental Samples
- Abbau von Pharmaka durch Biomasse aus Rückspülschlamm von Enteisenungs- und Entmanganungsfiltern eines Wasserwerks
- Optimisation of the Real-Time Polymerase Chain Reaction (qPCR) Method for the Quantification of Bacteriophages in Environmental Samples.
2017
- Wachstum von Eisenbakterien unter Huminsäure und verschiedenen Hefeextrakt-Konzentrationen. Bachelorarbeit.
- Untersuchung und Charakterisierung von Eisenbakterien auf mögliche Eisenreduktion, Kohlenstoff-Verwertung und Scheidenbildung. Praxisphasenbericht.
- Physiologische und phylogenetische Beschreibung von ausgewählten eisenoxidierenden Bakterie. Praxisphasenbericht.
- Vergleichende Genomanalysenund Bestimmung der Substratabbauspektren von Mangan und Eisenpräzipitierenden Bakterien. Bachelorarbeit.
2016
- Etablierung eines quantitativen Tests zur Bestimmung der Laccase Aktivität von Eisenbakterien. Bachelorarbeit.
- Einfluss von Pharmazeutika auf die initiale Adhäsion von Bakterien an Oberflächen. Bachelorarbeit.
- Weiterentwicklung und Validierung einer Extraktionsmethode zur getrennten Isolierung von eDNA und gDNA aus Biofilmen des γ - Proteobakteriums F8. Bachelorarbeit.
- Auswirkungen verschiedener Spurenstoffe auf das Wachstum von Eisenbakterien. Bachelorarbeit.
- Adaption of Iron and Manganese oxidizing Bacteria to treated Effluent. Bachelorarbeit.
- Untersuchungen von Eisenbakterien zur Toxizität und Biosorption von Schwermetallen. Bachelorarbeit.
2015
- Quantitativer Nachweis von Rhodoferax spp. in einer Wasserfassung mittels Real-Time PCR. Bachelorarbeit.
- Etablierung einer quantitativen Real-Time PCR zum spezifischen Nachweis von Crenothrix spp., Geothrix spp. Und Gallionella spp. in einer russischen Wasserfassung. Bachelorarbeit.
- Isolation of nitrifying bacteria adapted to Nitritox Fermenter. Masterarbeit.
- Untersuchungen zur kleinräumigen Heterogenitat von Bio lmpopulationen auf verockerten Unterwassermotorpumpen. Bachelorarbeit.
2014
- opulationsdynamik von Trinkwasserbiofilmen im kontinuierlich durchströmten Biofilmreaktor. Diplomarbeit.
- Vergleichende mikrobiologische Analysen von Reinwasserproben eines Wasserwerkes: ATP-Messung versus Durchflusszytometrie. Bachelorarbeit.
- Untersuchung zur Auswirkung von Wasch- und Reinigungsmitteln auf die EPS Struktur von Biofilmen mittels konfokaler Laser Scanning Mikroskopie. Bachelorarbeit.
- Entwicklung einer Extraktionsmethode zur getrennten Isolierung von eDNA und gDNA aus Biofilmen des g-Proteobakterium F8. Bachelorarbeit.
- Quantitativer Nachweis von Rhodoferax spp. in einer Wasserfassung mittels Real-Time PCR. Bachelorarbeit.
- Etablierung einer quantitativen Real-Time PCR zum spezifischen Nachweis von Crenothrix spp., Geothrix spp. und Gallionella spp. in einer russischen Wasserfassung. Bachelorarbeit.
2013
- Detection of a laccase genes and products in iron bacteria which degrade selected pharmaceutically active compounds. Masterarbeit
- Vergleich und Bewertung von vier Abwasserdesinfektionsverfahren hinsichtlich der Reduktion ausgewählter Mikroorganismen. Diplomarbeit.
- Charakterisierung von Bakterien aus einer russischen Wasserfassung. Bachelorarbeit.
- Anwendung der Probe Active Count (PAC)-Methode für neutrophile Eisen- und Mangan-ablagernde Bakterienstämme. Bachelorarbeit.
- Analyse von Biofouling auf Ultrafiltrationsmembranen. Diplomarbeit.
- Wirkungen von MVOC aus mikrobiell kontaminierten Gipskartonbauplatten auf Lungenzellen der Zelllinie A549. Diplomarbeit.
- Analyse der EPS Struktur von Biofouling bakterieller Reinkulturen auf Flachmembranen einer Labormembranfiltrationszelle mittels Confocal Laser Scanning Microscopy (CLSM). Diplomarbeit.
- Vergleichende Populationsanalysen der mikrobiellen Gemeinschaften einer russischen Wasserfassung. Bachelorarbeit.
- Impact of sediment desiccation and rewetting on benthic microbial diversity and carbon turnover. Diplomarbeit.
- Optimierung und Etablierung von Fluorochromen zur Charakterisierung von Biofouling auf Ultrafiltrationsmembranen. Bachelorarbeit.
- Interaktionen zwischen Aquabakterien und Pseudomonas aeruginosa in Trinkwasserbiofilmen. Diplomarbeit.
- Anwendung Fluoreszenz markierter Lektine zur Visualisierung von Polysacchariden in eiseninkrustierten Biofilmen. Studienarbeit.
- Charakterisierung von Bakterien aus einer russischen Wasserfassung. Bachelor.
- Vergleichende Populationsanalysen der mikrobiellen Gemeinschaften einer russischen Wasserfassung. Bachelorarbeit.
- Analyse von Biofouling auf Ultrafiltrationsmembranen. Diplomarbeit.
- Analyse der EPS Struktur von Biofouling bakterieller Reinkulturen auf Flachmembranen einer Labormembranfiltrationszelle mittels Confocal Laser Scanning Microscopy (CLSM). Diplomarbeit.
- Detection of a laccase genes and products in iron bacteria which degrade selected pharmaceutically active compounds. Masterarbeit.
2012
- Isolierung und Charakterisierung heterotropher Begleitbakterien von Cyanobakterien aus dem Berliner Müggelsee. Studienarbeit.
- Toxizitätsmessungen zur Bewertung des Risikos gewerblicher Abwässer für die kommunale Abwasserreinigung. Bachelorarbeit.
- Charakterisierung der EPS-Struktur von Biofouling eines Kläranlagenablaufs auf Hohlfasermembranen einer Crossflow-Membranfiltrationsanlage mittels konfokaler Laser Scanning Mikroskopie. Bachelorarbeit.
- Molekularbiologische Untersuchung eisenoxidierender mikrobieller Lebensgemeinschaften in Feuerland. Diplomarbeit.
- Optimierung von Fluoreszenzfärbungen von EPS Komponenten auf Ultrafiltrationsmembranen. Bachelorarbeit.
- Optimierung und Etablierung von Fluorchromen zur Charakterisierung von Biofouling auf Ultrafiltrationsmembranen. Bachelorarbeit.
- Fluoreszenzfärbung zur Charakteriesierung von Biofoulingkomponenten auf Ultrafiltrationsmembranen. Bachelorarbeit.
- Physiologische Untersuchung von eisenablagernden Betaproteobakterien. Bachelorarbeit.
- Consumption of Organic Nitrogen in Microbial Fuel Cells. Studienarbeit.
- Molekulare Charakterisierung von Cyanobakterienstämmen. Diplomarbeit.
- Development and Application of Specific Primers for Iron Precipitating Bacteria. Diplomarbeit.
- Abbau von Diclofenac mit Eisenoxidierenden Bakterien. Diplomarbeit.
2011
- Visualisierung der Biofilmstruktur auf Ultrafiltrationsmembranen. Diplomarbeit.
- Die Populationen von Pilzen in ruralen und urbanen Fließgewässern in Berlin und Brandenburg. Bachelorarbeit.
- Untersuchung des Wachstums methanogener Archaeen auf verschiedenen Marsregolith-Simulaten mit unterschiedlichen Fe(III)-Gehalt. Diplomarbeit.
- Vergleich des prozentualen Pilzgehalts aus den Holzarten Buche, Fichte, Kiefer und Eiche mittels DNA-Extraktion. Bachelorarbeit.
- Phylogenetische Untersuchung kultivierter neutrophiler Eisenbakterien aus dem Nationalpark Unteres Odertal. Bachelorarbeit.
- Mikrobiologischer Abbau von Paracetamol. Studienarbeit.
- Anreicherung von chemolithotrophen Eisenbakterien aus Feuerland in Gradientensystemen. Bachelorarbeit.
- Etablierung einer mikrokalorimetrischen Messmethode zur Untersuchung von Eisenbakterien. Studienarbeit.
- Untersuchung eisenoxidierender und -reduzierender Biofilme in Trinkwasserbrunnen. Bachelorarbeit.
- Biodegradation of Carbamazepine and Diclofenac by iron oxidizing bacteria. Projektarbeit.
- Evaluierung von Fluoreszenz markierten Lektinen zur Lokalisierung extrazellulärer polymerer Substanzen von verockerungsrelevanten Bakterien mittels Konfokaler Laser Scanning Mikroskopie. Bachelorarbeit.
- Vergleichende Populationsanalyse der laubassoziierten Pilze in ruralen und urbanen Fließgewässern. Diplomarbeit.
- Untersuchung des Abbaus von Huminstoffen durch Eisenbakterien. Bachelorarbeit.
- Untersuchungen zum Überleben und Wachstum von marinen Bakterien in Mars-relevanten Salzlösungen. Bachelorarbeit.
- Untersuchungen zum Wachstumsverhalten verockerungsrelevanter Bakterien aus Trinkwasserbrunnen. Diplomarbeit.
- Etablierung einer Oligonukleotid-Sonde zur Detektion
eisenablagernder Bakterien aus Trinkwasserbrunnen. Diplomarbeit. - Auswirkungen von extrazellulärer DNA in Bakterienbiofilmen auf den Fraßdruck durch Tetrahymena thermophila. Diplomarbeit.
- Detektion von verockerungsrelevanten Bakterien in Trinkwasserbrunnen. Bachelorarbeit.
2010
- Untersuchungen zur Austrocknungsresistenz von Eisenbakterien-Biofilmen. Bachelorarbeit.
- Morphologische und molekulare Taxonomie der aquatischen Pilzgattung Gyoerffyella. Studienarbeit.
- Molekularbiologische Untersuchungen von pathogenen Staphylococcus - Isolaten in Bezug auf Antibiotikaresistenzen, mobile genetische Elemente (MGEs) und Biofilmbildung. Diplomarbeit.
- Untersuchung der Diversität von Biofilmen in Trinkwasserbrunnen mittels Diversitäts-Assay und Denaturierender Gradienten Gel Elektrophore. Bachelorarbeit.
- Vergleich von Trinkwasserbiofilmen auf verschiedenen Materialien in einem Hausverteilungssystem mittels FISH und DGGE. Diplomarbeit.
- Isolierung und Charakterisierung von verockerungsrelevanten Mikroorganismen aus Trinkwasserbrunnen. Diplomarbeit.
- Morphologie eisenoxidierender Bakterien. Bachelorarbeit.
- Identifizierung von Antibiotika resistenten Milchsäurebakterien aus fermentierten Milch- und Fleischprodukten sowie molekulare Charakterisierung bezüglich Plasmidtransfer und Antibiotikaresistenzen. Bachelorarbeit.
- Untersuchungen zur Morphologie eisenoxidierender Grundwasserbakterien und ihrer Toleranz gegenüber Wasserstoffperoxid. Bachelorarbeit.
- Taxonomie der Gattung Heliscus basierend auf morphologischen und molekularen Merkmalen. Studienarbeit.
- Biotechnological and molecular characterisation of antibiotic resistant lactic acid bacteria isolated from dairy products. Studienarbeit.
- Phylogenetische Untersuchung von kultivierten Eisenbakterien aus Feuerland. Diplomarbeit.
- Populationsanalyse von Protozoen-Gesellschaften in Biofilmen mittels Fingerprinting- und Klonierungstechniken. Studienarbeit.
- Untersuchung der spezifischen DNA-Bindung verschiedener Transferproteine des konjugativen Plasmids pIP501 mittels Electrophoretic Mobility Shift Assay. Bachelorarbeit.
- Einfluss von Wasserstoffperoxid auf Biofilmpopulationen in Trinkwasserbrunnen. Diplomarbeit.
- Konjugativer Plasmidtransfer in Gram-positiven Bakterien: Studien zur DNA-Bindung und zum Plasmidtransfer zwischen Enterokokken und Staphylokokken. Bachelorarbeit.
- Nachweis und Lokalisierung von konjugativen Transferfaktoren in Gram-positiven Bakterien. Projekt MA.
- Isolierung und Charakterisierung heterotropher Begleitbakterien von Cyanobakterienkulturen. Studienarbeit.
- Influence of oxygen on a methanogenic consortium studied in a combined aerobic and anaerobic granular sludge reactor. Diplomarbeit.
- Use of Iron-Cycling Bacteria in Microbial Fuel Cells. Projektarbeit.
2009
- Untersuchung des Phenazonabbaupotentials eisenpräzipitierender Bakterien mittels LC-MS/MS. Bachelorarbeit.
- Wirksamkeit von Desinfektionsmitteln bei der Trinkwasseraufbereitung in Abhängigkeit vom pH-Wert:Untersuchungen an einer halbtechnischen Anlage. Projektarbeit.
- Molekularbiologische und biochemische Untersuchungen von Pilzbiozönosen in Fließgewässern. Diplomarbeit.
- Molekularbiologische Populationsanalyse von Trinkwasserbiofilmen auf verschiedenen Materialien. Diplomarbeit.
- Evaluierung von Oligonukleotid-Sonden zur Detektion von eisenpräzipitierenden Bakterien in nativen Bioflimen. Diplomarbeit.
- Populationsanalyse von Protozoen in Trinkwasserbiofilmen mit Hilfe von Fingerprinttechniken. Bachelorarbeit.
- Optimierung von Oligonukleotid-Sonden für den Nachweis von eisenpräzipitierenden Bakterien in nativen Bioflimen. Bachelorarbeit.
- Berechnung von FISH- Sonden für verblockungsrelevante Bakterien aus Trinkwasserbrunnen. Diplomarbeit.
- Vergleichende Populationsanylyse von Biofilmen des Unteren Odertals mittels DGGE. Projektarbeit.
- Reduktion suspendierter Biomasse durch bakterivore Mikroorganismen in sechs Naturnahen Systemen zur nachgeschalteten Abwasserbehandlung. Studienarbeit.
- Determination of phosphate and iron influence on the production of extracellular DNA. Diplomarbeit.
- Quantifizierung von extrazellulärer DNA. Projektarbeit.
- Vergleich und Bewertung mikrobiologischer, biochemischer und molekularbiologischer Quantifizierungsmethoden für Bakterien in Trinkwasserbiofilmen. Masterarbeit.
- Charakterisierung der Verockerung von Trinkwasserbrunnen. Masterarbeit.
- Studien zur Antibiotikaresistenz und zum horizontalen Gentransfer von Staphylococcus Isolaten der Internationalen Raumstation (ISS) und der Antarktis Forschungsstation CONCORDIA. Diplomarbeit.
- Untersuchung der spezifischen DNA-Bindung des putativen coupling-Proteins Orf10 des konjugativen Plasmids pIP501 mittels Electrophoretic Mobility Shift Assay. Praxisarbeit.
- Physiological characterization of iron and manganese precipitating bacteria from the National Park Unteres Odertal. Diplomarbeit.
- Specific detection of iron and manganese depositing Pedomicrobium spec. in biofilms of Nationalpark Unteres Odertal . Diplomarbeit.
- Untersuchung von gram-positiven bakteriellen Isolaten der CONCORDIA Forschungsstation auf Antibiotika-Resistenzen und Transfergene mit PCR. Praxisarbeit.
- Transformation und Markierung des aquatischen Pilzes Heliscus lugdunensis mit einem fluoreszierenden Protein. Diplomarbeit.
- Untersuchung des Estrogenabbaupotentials eisenpräzipitierender Bakterienstämme. Studienarbeit.
- Vergleichende Populationsanalyse von Bioflimen des Nationalparks Unteres Odertal. Studienarbeit.
- Charakterisierung von Bio lmpopulationen vor und nach einer Desinfektion mit der DGGE-Methode. Studienarbeit.
- Isolierung und Charakterisierung von Eisenbakterien aus Trinkwasserbrunnen. Diplomarbeit.
- Untersuchung der Wirksamkeit von Desinfektionsmitteln in der Trinkwasseraufbereitung mit F*-spezifischen Bakteriophagen an einer halbtechnischen Versuchsanlage. Diplomarbeit.
- Development of a novel high throughput continuous biotreatment process for dye wastewater. Diplomarbeit.
- Complete dechlorination of PCE by coupling of anaerobic and aerobic dechlorination in one reactor Studying the influence of carbon dioxide on the degradation of PCE by a methanogenic bioconsortium. Projektarbeit.
2008
- Klonierung des Replikationssystems des konjugativen Plasmids pIP501 in das Plasmid RP4 sowie Studien zur ATPase ORF10 und seiner ATPase Domäne. Masterarbeit.
- Charakterisierung von Bakterien aus marinen Sedimenten. Wissenschaftliche Abhandlung.
- Characterization of the methanogenic community structure in a laboratory scale biogas reactor using group specific real-time PCR. Diplomarbeit.
- Vergleich der Erfassungsbereiche von Isolierung und Klonierung anhand eines Bakterienmixes aus Gewässern des Unteren Odertals. Studienarbeit.
- Nachweis von Siderophorenproduktion mariner Mikroorganismen. Studienarbeit.
- Interspezifische Interaktionen zwischen Exiguobacterium acetylicum und eisenoxidierenden Pilzen aus dem Nationalpark „Unteres Odertal“. Diplomarbeit.
- Molekularbiologische Untersuchungen an bakteriellen Isolaten der Antarktisstation CONCORDIA und der Internationalen Raumstation ISS in bezug auf Biofilmbildung, konjugativen Transfer und Plasmidmobilisierung. Diplomarbeit.
- Kultivierung und molekularbiologische Analyse der mikrobiellen Biozönose von Sedimenten. Diplomarbeit.
- Vergleich der Pilzpopulationen von Blättern in urbanen und ruralen Fließgewässern während und nach dem Laubeintrag. Diplomarbeit.
- Development of Molecular Biological Methods for Detection of Amoebae in Drinking Water Biofilms. Diplomarbeit.
- Isolation and characterization of novel natural products from marine Actinomycetes. Studienarbeit.
- Optimierung der CARD-FISH-Methode für eisenoxidierende Bakterien im Biofilmen des Nationalparks "Unteres Odertal". Projektarbeit.
- Interaktionen zwischen Pilzen und Eisenbakterien aus der Mummert (Unteres Odertal). Diplomarbeit.
- Entwicklung der isothermen Titrationskalorimetrie zur Quantifizierung mikrobieller Abbauprozesse im Spurenbereich. Diplomarbeit.
- Taxonomie derr Gattung Heliscus basierend auf morphologischen und molekularen Merkmalen. Wissenschaftliche Abhandlung.
- Optimierung der FISH-Methode für eisenoxidierende Bakterien in Biofilmen des Nationalparks "Unteres Odertal". Projektarbeit.
- Optimierung sowie mikroskopische und phylogenetische Charakterisierung eines Biogasreaktors. Diplomarbeit.
- Analyse der Biofilme der Odert mittels Klonierung der 16S rDNA. Projektarbeit.
- Diversität von Biofilmgesellschaften im Nationalpark Unteres Odertal unter besonderer Berücksichtigung von Planctomyceten. Diplomarbeit.
- Entwicklung und Erprobung eines Anlagenkonzeptes zur Entfernung von Schwefelrückständen aus zur Biogasreinigung eingesetzten Biorieselbettreaktoren. Diplomarbeit.
- Klonierung und Expression des pIP501-codierten DNA-Sekretionsproteins ORF9 und Lokalisierung des Proteins in Enterococcus faecalis mit Immunoblot. Diplomarbeit.
- Physiologie und Phylogenie eisenoxidierender Bakterienisolate auf schadstoffhaltigen Medien. Diplomarbeit.
- Charakterisierung der bakteriellen Population aus den Rückspülschlämmen der Trinkwasseraufbereitung der Wasserwerke in Friedrichshagen, Kaulsdorf und Stolpe. Projektarbeit.
- Überprüfung der toxischen Wirkung auf Eukaryonten eines neuen von einem Urbanen Bakterienisolat produzierten Antibiotikums. Studienarbeit.
- Qualitativer und quantitativer Nachweis von Arcobacter sp.im Nachernteprozess der Spinatproduktion. Diplomarbeit.
- Quantifizierung von hygienisch relevanten Mikroorganismen in Trinkwasserbiofilmen. Diplomarbeit.
- Dispersion of culturable antibiotic resistant bacteria by flamingos in isolated and extreme environments of high altitude Andean wetlands. Projektarbeit.
- Intrazelluläre Lokalisierung des Transferproteins Orf7des konjugativen Plasmids pIP501 in Enterococcus faecalis sowie molekulare Untersuchungen zu Antibiotikaresistenzen von Bakterienisolaten der internationalen Raumstation ISS. Diplomarbeit.
- Studien zum Gentransfer in Biofilmen mit Staphylococcus- und Bacillus Isolaten der Internationalen Raumstation (ISS) und der Antarktisforschungsstation CONCORDIA. Bachelorarbeit.
- Klonierung und Expression des pIP501-kodierten DNA-Sekretionsproteins Orf6 sowie immunologische
Lokalisierung des Proteins in Zellkompartimenten von Enterococcus faecalis mittels Immunoblot. Studienarbeit. - Isolation and Characterization of Biosurfactant-Producing Marine Bacteria from Izmir Inner Bay, Aegean Sea. Diplomarbeit.
2007
- Diversität von Biofilmgesellschaften in Gewässern im Nationalpark Unteres Odertal unter besonderer Berücksichtigung von eisen- und manganoxidierenden Bakterien. Diplomarbeit.
- Klonierung, Expression, Reinigung und enzymatische Charakterisierung des Transferproteins ORF10 aus dem Plasmid pIP501. Diplomarbeit.
- Diversity of Archaea from a hot spring in Pisciarelli Solfatara determined by molecular and cultural methods. Projektarbeit.
- Studien zur bakteriellen Diveristät und zu Antibiotika-Resistenzen in Wasser- und Vogelkotproben aus hochgelegenen andinen Feuchtgebieten. Studienarbeit.
- In situ Studien der Protein-Protein-Interaktionen der zwei Domänen CHAP und SLT des Transferproteins Orf7 mit den Transferproteinen Orf5, Orf7, Orf10 und Orf14 des konjugativen Plasmids pIP501 aus Streptococcus agalactiae. Diplomarbeit.
- Weiterführende Optimierung der CARD-FISH-Methode zur Detektion von Bodenbakterien aus unterschiedlichen urbanen Habitaten. Projektarbeit.
- Neue Protein-Protein-Interaktionen zwischen Virulenzfaktoren von Enterococcus faecalis V583 und humanen Proteinen. Studienarbeit.
- Aktivitätsbestimmung mittels CTC an Escherichia coli aus einem Membranreaktor. Studienarbeit
- Vitalitätsbestimmung eines Escheria Coli Umweltisolats in einem Membranbioreaktor während einer Langzeituntersuchung. Studienarbeit.
- Interaktion zwischen Pilzen und Bakterien: Nachweis und räumliche Untersuchung von Bakterien in der Hydrosphäre von Pilzen mit SYTO®9 und DGGE von Pilzpopulationen auf Blättern aus urbanen Bächen. Studienarbeit.
2006
- Morphologische und physiologische Untersuchung an nicht-wachsenden Escherichia coli - Zellen aus einem Membranbioreaktor. Projektarbeit.
- Bakterien aus marinen Sedimenten als Produzenten antifungaler Sekundärmetabolite. Diplomarbeit
- Charakterisierung der bakteriellen Bodenbiofilmpopulation und Optimierung der CARD-FISH-Methode zur Detektion von Bodenbakterien. Diplomarbeit.
- Überprüfung der EPS Produktion von drei bakteriellen Biofilmisolaten in Abhängigkeit vom Wachstumsmmedium. Projektarbeit.
- Development of a coupled methanogenic/methanotrophic biofilm consortium for complete degradation of tetrachloroethylene and study of its microbial ecology. Diplomarbeit.
- Vergleichende Untersuchungen zum Nachweis von Polyphosphaten in Belebtschlämmen. Diplomarbeit.
- Mikrobiologische Charakterisierung neuartiger mariner Mikroorganismen. Diplomarbeit.
- Interactions between Fungi on willow leaves in an aquatic microcosm. Diplomarbeit.
- Prüfung der Wirksamkeit von Desinfektionsmitteln für das Wasserwerk sowie für die Hausinstallation mit Hilfe von Biofilmkulturen. Diplomarbeit.
- Taxonomie der Gattung Alatospora basierend auf mophologischen und molekularen Merkmalen. Diplomarbeit.
- Isolierung und Charakterisierung von Bodenbakterien nach einer Austrockungszeit von drei Monaten. Studienarbeit.
- Molekularbiologische und physiologische Untersuchungen von Escherichia coli im Erhaltungsstoffwechsel während der Langzeitkultivierung in einem Membranbioreaktor. Diplomarbeit.
- Entwicklung eines computergestützten Systems zur automatisierten Morphometrie von Bakterien
2005
- [A] Bindungsstudien der TraA-Relaxase und der N-terminalen Domäne TraAN246 am oriT des Plasmids pIP501 anhand von footprint Analysen. [B] Klonierung und Expression der TraA-Relaxase und der beiden N-terminalen TraAN246- und TraAN269-Domänen des Plasmids pIP501. Studienarbeit.
- Klonierung, Expression und Charakterisierung der von orf7 des konjugativen Plasmids pIP501 codierten putativen lytischen Transglykosylase. Diplomarbeit.
- Charakterisierung der bakteriellen Bodenbiofilmpopulation und Optimierung der CARD-FISH-Methode zur Detektion von Bodenbakterien. Diplomarbeit.
- Untersuchung der Physiologie des Stammes E. coli A3 in der stationären Phase. Diplomarbeit.
- Protein-protein interaction studies of the pIP501 transfer genes orf1, orf8 and orf15 with the yeast two-hybrid system. Projektarbeit.
- In vitro Studien der Protein-Protein-Interaktionen der Transferproteine Orf5 und Orf7 des konjugativen Plasmids pIP501 aus Streptococcus agalactiae. Diplomarbeit.
- Klonierung und Expression der putativen Gentransferproteine ORF8 und ORF15 des Multiresistenzplasmids pIP501 in den His tag Vektor pQTEV mit anschließender Reinigung der Fusionsproteine durch Affinitätschromatographie. Diplomarbeit.
- Isolierung und Charakterisierung von wachsabbauenden Mikroorganismen zur Reduktion der Hydrophobie von Böden. Projektarbeit.
- Einfluss von Biofilmen hydrophober und hydrophiler Bakterienisolaten urbaner Standorte auf die Benetzbarkeit von Bodenproben. Diplomarbeit.
- Charakterisierung der bakteriellen Biofilmgesellschaften in einem Langsamsandfilter. Diplomarbeit.
- Vergleich und Charakterisierung der extrazellulären Filamente des gamma-Proteobakteriums F8 und zwei weiterer unbekannter Bakterienstämme. Diplomarbeit.
- Versuche zur Klonierung der Mobilisierungsregion und zweier Transfergene des konjugativen Plasmids pIP501 in einen E. coli / E. faecalis Shuttle Vektor. Projektarbeit.
- Einfluss von Wasserstress auf die bakterielle Gemeinschaft eines urbanen Bodens. Bachelorarbeit
- Studies on protein-protein interactions of Enterococcus faecalis virulence proteins with human proteins by the yeast two-hybrid system. Diplomarbeit.
- Untersuchungen zum Abbau von Phenazon in Bereichen erhöhter bakterieller Eisenoxidation bei der Trinkwasseraufbereitung. Diplomarbeit.
- Klonierung und Expression des vom konjugativen Plasmid pIP501 kodierten Transferproteins ORF5 in Escherichia coli. Studienarbeit.
2004
- Isolierung von Mikroorganismen aus urbanen Böden und Untersuchung ihrer Abbauleistung von Phenanthren. Projektarbeit.
- Biofilme des Auengebietes "Unteres Odertal" und deren Beeinflussung durch Arzneimittel. Diplomarbeit.
- Characterization of unusual microfilaments of an hitherto unknown gamma-Proteobacterium. Projektarbeit.
- Limnologische Untersuchung: "La Laguna del Tejo de Canada del Hoyo". Projektarbeit.
- Untersuchung der extrazellulären Mikrofilament-DNS des gamma-Proteobakteriums F8. Projektarbeit.
- Untersuchungen zur Einsetzbarkeit des CHEMSCAN®RDI-Laserzytometers zum Nachweis von Mikroorganismen aus Umweltproben. Projektarbeit.
- Krankheitserreger im Trinkwasser. Studienarbeit.
- Vergleich unterschiedlicher Methoden zur Bestimmung der Biomasseentwicklung eines Escherichia coli Umweltisolats in zwei verschiedenen Langzeitkultivierungsverfahren. Studienarbeit.
- Physiologie, Kultivierbarkeit und automatische Bildanalyse von Escherichia coli in Langzeituntersuchungen. Studienarbeit.
2003
- Protocol for generating axenic Ulva lactuca and studies of type IV pili mediated attachment of Pseudoalteromonas tunicata to the algal surface. Diplomarbeit.
2002
- Untersuchungen zur Besiedlung von Oberflächen durch Mikro- und Makrofauna. Diplomarbeit.
- Trinkwasseraufbereitung und deren Einfluss auf die mikrobiologische Qualität. Projektarbeit im Rahmen des Umwelttechnischen Seminars.
- Erfassung mikrobiologischer Parameter in einem rechnergesteuerten Membranreaktor. Diplomarbeit.
- Untersuchungen der Regulation des konjugativen Transfers in pathogenen Mikroorganismen mit hygienischer Bedeutung für Lebensmittel am Beispiel des Plasmids pIP501. Diplomarbeit.
- Phylogenetische Einordnung der Pilze Anguillospora longissima und A. crassa aufgrund ribosomaler Gensequenzen. Examensarbeit.
- Phylogenetische Analyse von 18S- und ITS-Sequenzbereichen der aquatischen Pilze Tricladium splendens und Tricladium angulatum. Examensarbeit.
2001
- Expressionsstudien der Transfergene des Plasmids pIP501 anhand des Reportergens gfp sowie GFP-Detektion mittels Epifluoreszenzmikroskopie in Streptococcus pneumoniae und Bacillus subtilis. Staatsexamensarbeit.
- Evaluation of the degree of correlation of experimental respiratory methods (our) and the activity of the enzyme dehydrogenase (DHA) to determine the decay coefficient of heterotrophic biomass (bH). Diplomarbeit.
- Mikrobiologische und hygienische Aspekte beim Einsatz von EPDM-Gummischläuchen in Trinkwasserinstallationen. Diplomarbeit.
- Bestimmung des Transkriptionsstarts der tra mRNA des konjugativen Plasmids pIP501 sowie DNA Sequenzbestimmung eines weiteren Transfergens. Staatsexamensarbeit.
- Toxizität und Microcystingehalt von Extrakten einer Planktothrix agardhii-Freilandblüte und zweier Kulturstämme. Diplomarbeit.
- Characterization of biofilms from Australian drinking water distribution systems. Diplomarbeit.
2000
- Mikrobiologische Untersuchungen zur Struktur und Funktion eines nitrifizierenden Rotationsscheibenreaktor-Biofilms. Studienarbeit.
1999
- Phylogenetische Diversität und räumliche Verteilung von mesophilen, Gram-negativen sulfatreduzierenden Bakterien in einem anaeroben Festbettreaktor. Studienarbeit.
- Untersuchung homoacetogenen Mikroorganismen in einem anaeroben Festbettreaktor. Diplomarbeit.
- Untersuchungen der physiologischen Aktivität von Belebtschlämmen in einer Abwasserreinigungsanlage mit integrierter Mikrofiltration. Studienarbeit.
- Transformation of Reactive Violet 5 by the white-rot fungus Bjerkandera adusta - characterization of the metabolite patterns. Diplomarbeit.
- Mikrobiologische Untersuchungen an hochkonzentriertem Belebtschlamm aus einer membrangestützen Mikrofiltration zur Reinigung kommunaler Abwässer. Diplomarbeit.
1998
- Populationsanalyse einer reduktiv dechlorierenden Mischkultur. Studienarbeit.
- Isolation und Charakterisierung aerober Mikroorganismen aus dem marinen Schwamm Chondrosia reniformis. Diplomarbeit.
- Isolierung und Charakterisierung einiger Vertreter einer anaerob Trichlorbenzole dechlorierenden Bakterienmischkultur. Diplomarbeit.
1997
- Transformation von Remazol Brillantviolett 5R durch Bjerkandera adusta - physiologische und enzymatische Untersuchungen. Diplomarbeit.
- Isolierung und Charakterisierung von anaeroben Mikroorganismen aus einer Mischpopulation in dem marinen Schwamm Chondrosia reniformis. Diplomarbeit.
1996
- Wechselwirkungen zwischen der Bodenpilzflora, dem pH-Wert und der Freisetzung von PCB 52 in einem Rieselfeldboden. Diplomarbeit.
1995
- In situ Untersuchungen von anaeroben Prokaryonten im Belebtschlamm mit rRNS-gerichteten, fluoreszenzmarkierten Oligonukleotidsonden. Studienarbeit.
Bewertungskriterien und Anleitung
Bewertungskriterien und Anleitung
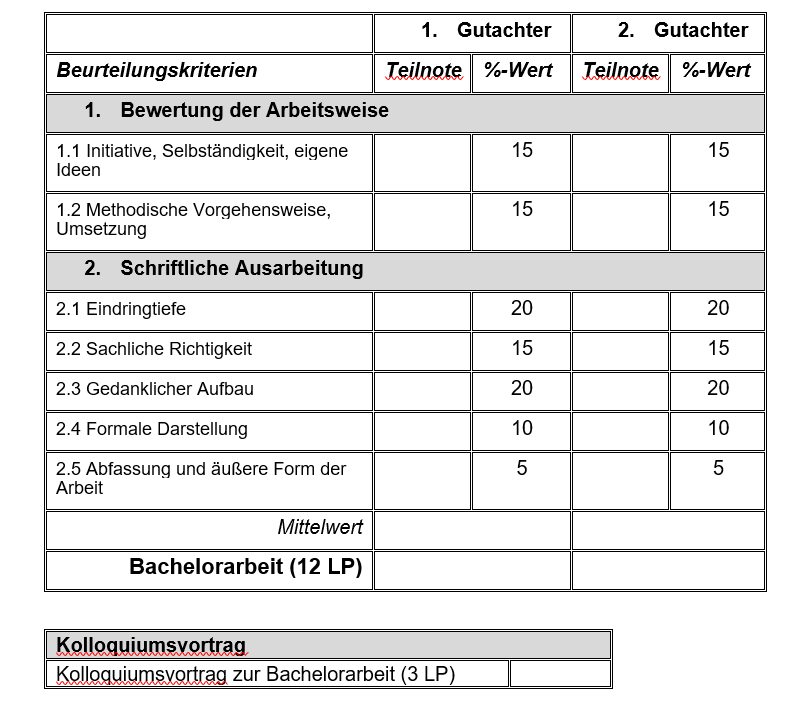
Zur Bewertung von Bachelor-, Master-, Studien- und Diplomarbeiten werden an unserem Fachgebiet die in Tabelle 1 aufgelisteten Kriterien herangezogen. Bei jedem einzelnen Kriterium müssen mindestens 25 % der Punkte erreicht werden. In Tabelle 2 finden Sie den Schlüssel, um die erreichte Prozentzahl in eine Note umzurechnen.
Tabelle 2: Notenschlüssel
| Note | Prozentzahl |
|---|---|
| 1,0 | ab 95% |
| 1,3 | ab 90% |
| 1,7 | ab 85% |
| 2,0 | ab 80% |
| 2,3 | ab 75% |
| 2,7 | ab 70% |
| 3,0 | ab 65% |
| 3,3 | ab 60% |
| 3,7 | ab 55 % |
| 4,0 | ab 50% |
| 5,0 | 0% |
Standort
Kontakt
| Sekretariat | BH 6-1 |
|---|---|
| Gebäude | Bergbau Hütten Neubau |
| Adresse | Ernst-Reuter-Platz 1 10587 Berlin |

